Proteins are fundamental to all biological processes, playing critical roles in growth, metabolism, and various cellular functions. Each protein’s structure directly influences its functionality, a concept that is vital in biochemistry. If the structure of a protein is altered, even slightly, it can lead to dysfunction and potentially serious health issues. Despite this general rule, a fascinating category of proteins defies fixed structural norms. The study of these flexible proteins is gaining traction within the scientific community, particularly as researchers uncover new methods to study them in more natural settings.
Recently, researchers from Martin Luther University Halle-Wittenberg and the National Hellenic Research Center have made significant strides in understanding one such protein: myo-inositol-1-phosphate synthase (MIPS). This protein is integral to the biosynthesis of inositol, often referred to as vitamin B8, which, in addition to its metabolic roles, plays part in cellular signaling and overall health. MIPS is not unique in its importance; however, its behavior as an intrinsically disordered protein presents intriguing questions about protein dynamics.
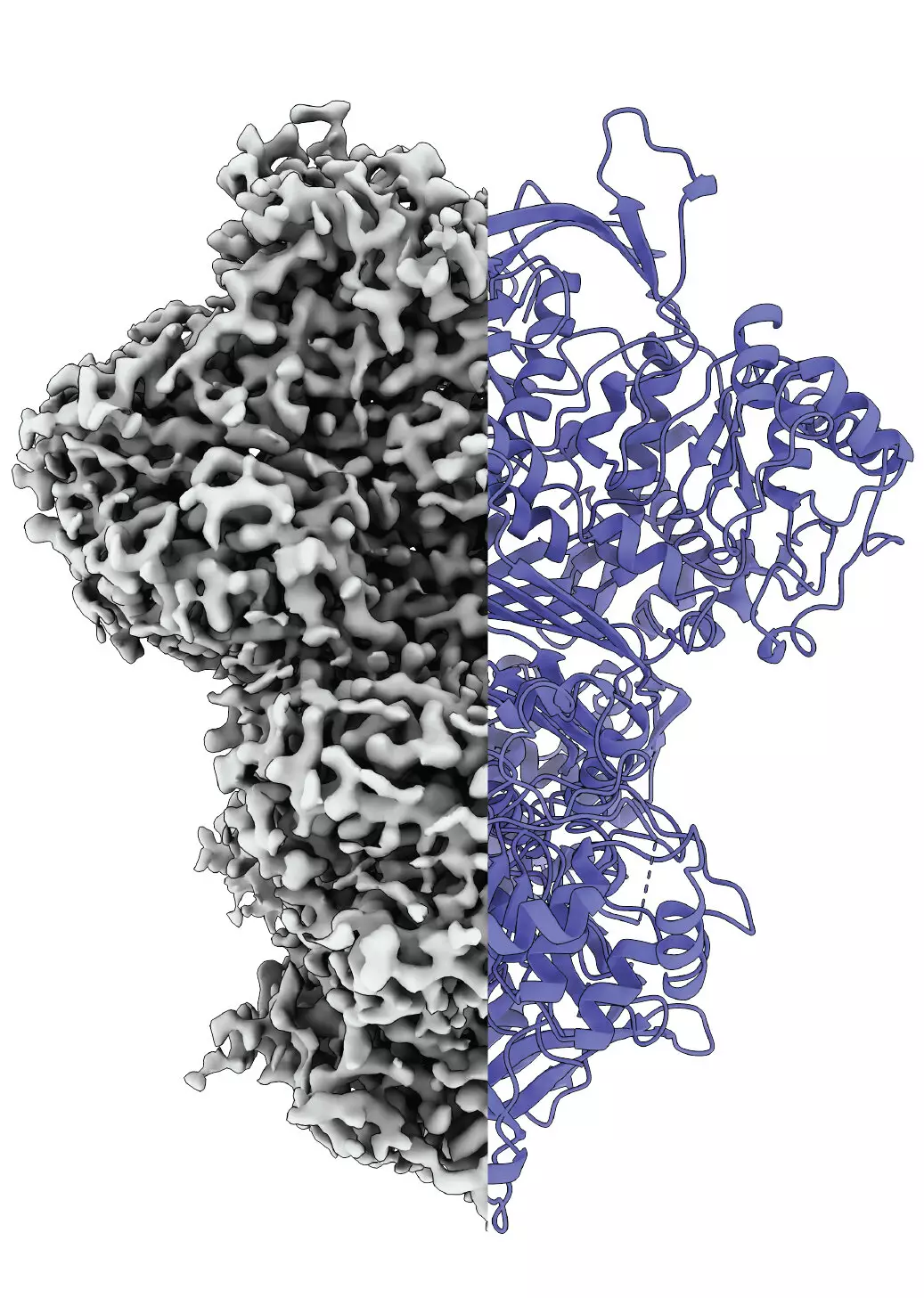
In a groundbreaking study reported in the Proceedings of the National Academy of Sciences, the MIPS team utilized cryo-electron microscopy to visualize the protein in action. This approach allowed them to observe MIPS transitioning between different structural states: a disordered form, an ordered conformation, and an intriguing intermediate state. The existence of this third state raises questions regarding its functional significance. Understanding why MIPS adopts such diverse conformations could illuminate fundamental biochemical mechanisms—perhaps it assists in molecular interactions related to hydration or serves another unknown purpose.
What sets this research apart is the innovative methodology employed by the team. Traditional protein analysis often strips proteins from their natural environments, limiting insights into their dynamic behavior. However, Professor Panagiotis Kastritis and his group have developed techniques to observe proteins under conditions that closely mimic their physiological states. This methodological advancement could usher in a new era of protein research, expanding possibilities for discovering how various proteins perform their essential biological roles.
MIPS belongs to a broader family of proteins known as isomerases. As this research delves deeper, the Kastritis team has begun exploring whether other isomerases display similar dynamic behavior to MIPS. Analyzing over 340 related proteins, they found evidence suggesting that the fluctuations in structure may be a common trait among these enzymes, challenging the notion that protein stability is essential for function.
This realization has profound implications not only for basic science but also for the development of therapeutic strategies. By enhancing our understanding of metabolic pathways—how proteins interact and change shape throughout biochemical processes—scientists like Kastritis may be laying the groundwork for novel treatments targeting diseases linked to dysfunctional proteins. The research highlights the importance of not merely isolating proteins for study but integrating their environmental contexts into the analysis.
The work conducted on MIPS exemplifies the evolution of protein research, emphasizing the notion that flexibility and adaptability may be inherent features of many enzymes. As researchers continue to unravel the complexities of protein dynamics, the potential for breakthroughs in understanding metabolic disorders and therapeutic development grows. The findings from Halle-Wittenberg and the National Hellenic Research Center showcase the power of innovative research techniques and collaborative efforts, paving the way for compelling new discoveries in the exciting field of biochemistry. As we learn more about the dynamic world of proteins, we may soon unlock answers that redefine our understanding of life at the molecular level.

Leave a Reply